Authored by Tsitsiashvili Gurami*
Introduction
By analogy with [1], we consider a protein network represented by a directed graph (digraph) G with the set U of nodes, which are proteins and whose directed edges are paired bonds between nodes represented in the Cytoscape program. Dedicate the subset U/ ⊆U of nodes and decrease in a comparison with [1] a number of edges, which block all paths from outside of the set U/ outside of the setU/ .
Take all nodes from the subset U/ and all edges between them. These nodes and edges create directed sub-graphG/ ⊆ G . Replace all (directed) edges of the sub-graph G/ by undirected ones and obtain undirected graph G// . Define in the sub-graph G// all its connectivity components / / / / Return directions to all edges of the subgraph G//k G k and define in such a way the sub-graph Gk of the digraph G, k =1,...,n.
Factorize each sub-graph Gk by a relation of cyclic equivalence (two nodes are cyclically equivalent if there is a cycle containing both of them) and construct acyclic digraph with nodes - clusters of cyclic equivalence. Construct the partial order ≥ between the clusters (a relation A ≥ B is true if there is a way from cluster A to cluster B ) in Gk .
Denote by vk *a set of edges incoming to GK and by VK**a set
of edges out-coming from GK and designate bypK, qKnumbers of
edges in these sets. Define the following sets WK*,WK**of clusters
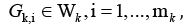 in digraphGK Each cluster
in digraphGK Each cluster
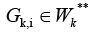 has
final nodes of some edges from the set V
K*Similarly, each cluster
has
final nodes of some edges from the set V
K*Similarly, each cluster 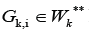 has initial nodes of some edges from the set VK
**The WK*,WK** setsmay intersect.
has initial nodes of some edges from the set VK
**The WK*,WK** setsmay intersect.
Construct the set 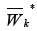 consisting of clusters GK,i,such that
consisting of clusters GK,i,such that
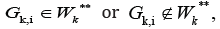 but there are some cluster
but there are some cluster
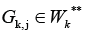 and a way from
and a way from
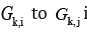 in digraph GK.By analogy construct the set
in digraph GK.By analogy construct the set
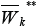 of clusters
of clusters
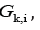 such that
such that
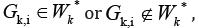 but
there are some cluster
but
there are some cluster 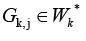 and a way from
and a way from
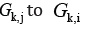 in
digraph GK
in
digraph GK
Designate by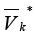 all edges incoming some clusters containing
in the set
all edges incoming some clusters containing
in the set 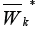 from the outside of the graph
from the outside of the graph
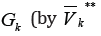 all edges
out-coming from some clusters in the set WK**outside the graph GKDenote
all edges
out-coming from some clusters in the set WK**outside the graph GKDenote
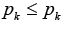 a number of edges in the set
a number of edges in the set
 a number of edges in the set
a number of edges in the set
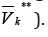
Assume that 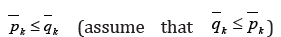 and block
all edges from
and block
all edges from  In such a way,
we block all paths from the outside of GKoutsideGK. Then a number of edges blocking the sub-graph GKequals’
In such a way,
we block all paths from the outside of GKoutsideGK. Then a number of edges blocking the sub-graph GKequals’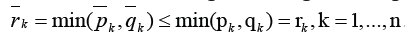 Last
inequalities
show, how this algorithm decreases a number of blocked edges in a
comparison with [1]. Next step of the decomposition procedure in
this algorithm may be an allocation of disconnected subsets in pairs
of sets
Last
inequalities
show, how this algorithm decreases a number of blocked edges in a
comparison with [1]. Next step of the decomposition procedure in
this algorithm may be an allocation of disconnected subsets in pairs
of sets 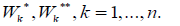
To read more about this article...Open access Journal of Biostatistics & Biometric Applications
Please follow the URL to access more information about this article
https://irispublishers.com/abba/fulltext/decomposition-algorithms-of-blocking-the-selected-edges-in-the-digraph.ID.000562.php
To know more about our Journals....Iris Publishers





No comments:
Post a Comment