Authored by Ricardo Gobato*,
Mini Review
The Deoxyribonucleic acid (DNA) is a molecule composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. The A, C, G, and T, - adenine, cytosine, guanine and thymine, respectively, representing the four nucleotide bases of a DNA strand - adenine, cytosine, guanine, thymine-covalently linked to a phosphodiester backbone.
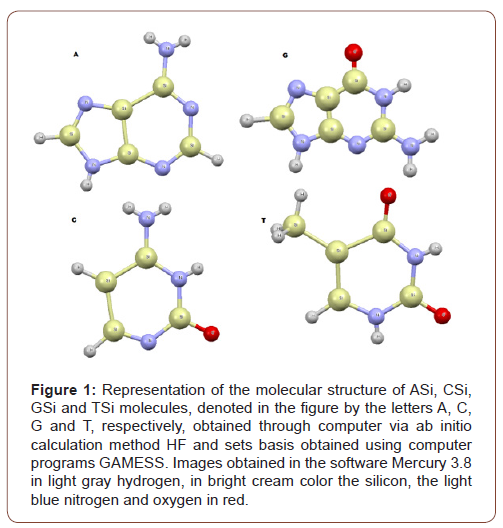
The core of the work is based on the replacement of carbon atoms by silicon atoms, on the basis of four standard bases of DNA: A, C, G and T (adenine, cytosine, guanine, thymine). Determining with minimum computational methods via ab initio Hartree-Fock methods, their minimum formation energies, specific heat, entropy, infrared spectrum and their peak absorbance frequencies.
The option for simple replacement of carbon by silicon is due to the peculiar characteristics between both. Atomic interactions under non-carbon conditions were studied, with only the Hydrogen, Silicon, Nitrogen and Oxygen atoms, in STP (Standard Temperature and Pressure), for the four standard bases of DNA, A, C, G and T, thus obtaining by quantum chemistry four new compounds, named here as: ASi, CSi, GSi and TSi.
In comparing the carbon and silicon has: the Si lies in the same column of the periodic table of the elements, and t has been investigated as a possible alternative for building up biological molecules in exobiology. Silicon based chemistry, however, is by far less flexible than carbon chemistry, not able to form double covalent bonds with the same easiness as Carbon does. Other fact is the larger volume occupied by the external electronic orbitals of silicon tend to reduce the superposition of p orbitals.
Through the chemical abundances of biological elements in the earth crust, terrestrial life has chosen carbon instead of silicon, in spite of the larger abundance of silicon. This fact suggests that carbon is better suited to form biological molecules.
However, this paper assumes conditions without the presence of Carbon. The molecular structure of ASi, CSi, GSi and TSi molecules, were obtained through computationally calculated molecular dynamics, using the ab initio RHF (Restrict Hartree-Fock method) and UHF (Unrestricted Hartree-Fock method). The set of basis used that have are accepted by quantum chemistry parameters, with ab initio methods, in the bases CC-pVTZ (Correlation-consistent valence-only basis sets triple-zeta) and 6-311G**(3df, 3pd), UHF CEP-31G (ECP (Effective core potential (minimal basis) split valance) and UHF CEP-121G (ECP triple-split basis).
The ab initio calculations have been performed to study the equilibrium configuration the molecules using the GAMESS. The names of the new molecules obtained ASi, CSi, GSi and TSi, are: 2,3,4,5,6,7,8,9-octahydro-1H-[1,3,2,4,5]diazatrisilolo[4,5-d] [1,3,2,4,5,6]diazatetrasilin-8-amine; 2-hydroxy-1,3,2,4,5,6- diazatetrasilinan-4-amine; 8-oxo-3,7-dihydro-[1,3,2,4,5] diazatrisilolo[4,5-d][1,3,2,4,5,6]diazatetrasilin-6-amine and (2,4-dihydroxy-1,3,2,4,5,6-diazatetrasilinan-5-yl) silane, respectively.
Studies did not reveal any works with characteristics studied here. There is an absence of a referential of the theme, finding only one work in (Gobato et al., 2018, 2019, 2020, 2021). The molecular Hartree-Fock wave function is written as an ant symmetrized product (Slater determinant) of spin-orbitals, each spin-orbital being a product of a spatial orbital ϕi and a spin function (either α or β).
The expression for the Hartree-Fock molecular electronic energy E(HF )is given by the variation theorem as where D is the Slater-determinant Hartree-Fock wave function and 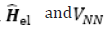 are given by
are given by
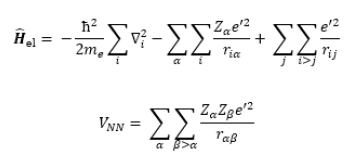
Since V_NN does not involve electronic coordinates and D is normalized, we have
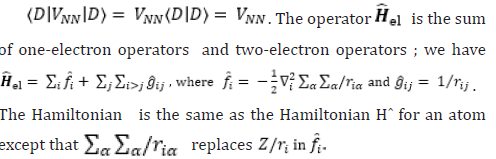
To read more about this article....Open access Journal of Biomedical Engineering & Biotechnology
Please follow the URL to access more information about this article
To know more about our Journals...Iris Publishers





No comments:
Post a Comment