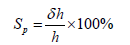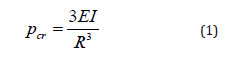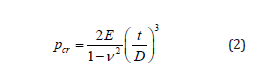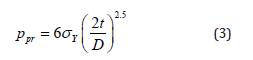Authored by Farid E Ahmed*
Introduction
Colorectal cancer (CRC) is the third most common malignancy
worldwide, with an estimated one million new cases and half
million deaths yearly. Screening for CRC allows early stage
diagnosis of malignancy and potentially reduces disease mortality.
The convenient and inexpensive fecal occult blood test (FOBT)
screening test has low sensitivity and requires dietary restriction,
which impedes compliance. Although colonoscopy. Is the golden
screening standard for the for this cancer, the invasive nature,
abdominal pain and high cost have hampered worldwide application
of this procedure. A noninvasive sensitive screen for colon cancer
(CC) requiring no dietary restriction is a more convenient test. CC is
more abundant in the USA than rectal cancer (RC).
The discovery of small non-coding protein sequences, 17-
27 nucleotides long RNAs (such as microRNAs), has opened new
opportunities for a non-invasive test for early diagnosis of many
cancers. MiRNA functions seem to regulate development and
apoptosis, and specific miRNAs are critical in oncogenesis, effective
in classifying solid and liquid tumors, and serve as oncogenes or
suppressor genes. MiRNA genes are frequently located at fragile
sites, as well as minimal regions of loss of heterozygosity, or
amplification of common break-point regions, suggesting their
involvement in carcinogenesis. Profiles of miRNA expression differ
between normal tissues and tumor types, and evidence suggests
that miRNA expression profiles can cluster similar tumor types
together more accurately than expression profiles of protein-coding
mRNA genes. Although exosomal RNA are missed, a parallel carried
out on stool miRNAs to compare the extent of loss when colonocytes
are only used can be carried out, and an appropriate corrections for
exsosomal loss can be made. To ascertain the validity of a miRNA
screening test for CC, it must be validated in a study, using a nested
case control epidemiology design and employing a prospective
specimen collection, retrospective blind evaluation (PRoBE) of
control subjects and test colon cancer patients, as delineated by
NCI’s Early Detection Research Network (EDRN) http://edrn.nci.
nih.gov. Immunoparamagnetic are employed to capture colonocytes
from harsh stool environment, whose extracted fragile total small
RNA is stabilized shortly after stool excretion by commercial kits
so it does not ever fragment, followed by standardized analytical
quantitative miRNA dPCR-chip profiling in noninvasive stool
samples, to develop a panel of few stable miRNAs for absolute
quantitative diagnostic screening of early sporadic colon cancer
(stage 0-1), more economically and with higher sensitivity and
specificity than other CC screening test on the market today.
A preliminary global microarray expression analysis using
an exfoliated colonocytes enrichment strategy, which employed
control subjects and various stages (0-4) of CC, using Affymetrix
Gene Chip miRNA 2.0 Array, showed 180 preferentially expressed
miRNA genes that were either increased (124 miRNAs), or reduced
(56 miRNAs) in expression in stool samples from CC patients. This
allowed careful selection of 14 miRNAs (12 Up-Regulated, miR-19a,
miR-20a, miR-21, miR-31, miR-34a, miR-96, miR-106a, miR-133a,
miR-135b, miR-206, miR-224 and miR-302; and 2 Down-Regulated,
miR-143 and miR-145) Table 1 for further PCR analysis (Table 1).
Then analysis carried out using absolute miRNAs expression by
a chip-based digital PCR by partitioning a sample of DNA or cDNA
into many individual, parallel PCR reactions; some of which contain
the target molecule (positive), while others do not (negative). A
single molecule can be amplified a million-fold or more. During
amplification, TaqMan chemistry with dye-labeled probes is used
to detect sequence-specific targets. When no target sequence
is present, no signal accumulates. Following dPCR analysis, the
fraction of negative reactions is used to generate an absolute count
of the number of target molecules in the sample, without the need
for standards or endogenous controls. In conventional qPCR, the
signal from wild-type sequences dominates and obscures the
signal from rare sequences. By minimizing the effect of competition
between targets, dPCR overcomes the difficulties inherent to
amplifying rare sequences and allows for sensitive & precise
absolute quantification of the selected miRNAs. Applied Biosystem
Quant Studio™ 3D instrument only performs the imaging and
primary analysis of the digital chips. The chips themselves must be
cycled offline on a Dual Flat Block Gene Amp® 9700 PCR System or
the ProFlex™ 2x Flat PCR System. The Quant Studio™ 3D Digital PCR
System (Figure 1) can read the digital chip in less than 1 minute,
following thermal cycling (Figure 1).
1. Chip Case Lid- The lid used to seal the Digital PCR 20K
Chip for thermal cycling and imaging on the Quant StudioTM
3D Instrument.
2. Digital PCR 20K Chip- The 10-mm2 consumable that
contains the 20,000 reaction wells, which suspend the
individual PCR reactions for thermal cycling and imaging.
3. Quant StudioTM 3D Digital PCR Chip Case- The thermal
-conductive base that secures and protects the Digital PCR 20K
Chip during all phases of use.
4. Chip ID- A label applies to the Quant StudioTM 3D Digital
PCR chip Case Lid that can be used to uniquely identify the chip
to which it is applied.
5. Fill Port- The aperture within the Chip Case Lid through
which immersion Fluid is injected on to the Chip.
6. Reaction Wells- The 20,000 physical holes within the
Digital PCR 20K Chip that suspend the individual PCR reaction.
The current Quant Studio™ 3D Digital PCR Chip allows for
one sample per chip; although, duplexing allows for analysis of
two targets per chip. Sample prep for digital PCR is no different
than for real-time PCR, when using the Quant Studio™ 3D Digital
PCR System. The concentration of cDNA stock can be estimated
by including all of the necessary dilution factors into the Analysis
Suite™ software, which gives the copies/μL in the stock. A critical
step in dPCR, is sample partitioning [i.e., division of each sample
into thousands of discrete subunits prior to amplification by PCR,
each ideally containing either zero or one (or at most, a few)
template molecules]. Each partition behaves as an individual
PCR reaction –as with real-time PCR—fluorescent FAM probes
[or others, as VIC fluorescence. Samples containing amplified
products are considered positive (1, fluorescence), and those
without product –with little or no fluorescence are negative (0,
fluorescence). The ratio of positives to negatives in each sample
is the basis of amplification. Unlike real-time qPCR, dPCR does
not rely on the number of amplification cycles to determine the
initial amount of template nucleic acid in each sample, but it relies
on Poisson Statistics to determine the absolute template quantity.
The unique sample partitioning step of dPCR, coupled with Poisson
Statistics, allows for higher precision than both traditional and
qPCR methods; permitting for analysis of rare miRNA targets. The
use of a nanofluidic chip provides a convenient mechanism to run
thousands of PCR reactions in parallel. Each well is loaded with a
mixture of sample, master mix, and Applied Biosystems TaqMan
Assay reagents are individually analyzed to detect the presence
(positive) or absence (negative) of an endpoint signal. To account
for wells that may have received more than one molecule of the
target sequence, a correction factor is applied using the Poisson
model. It features a filter set that is optimized for the FAM™, VIC®,
and ROX™ dyes, available from Life Technologies.
Absolute quantification of the 14 miRNAs is shown in Table 2,
and Table 3 is a representation of SDs and R2 for the 14 miRNAs
tested by absolute digital PCR. (Figure 2) is Workflow of a digital
miRNA’s PCR for colon cancer profiling in human colon tissue or
stool samples. (Figure 3). is a graphical representation of the
absolute quantification of the 12 up- or 2 down-regulated miRNAs
in Human Stool by the QuantStudioTM 3D Digital PCR Chip System.
Digital PCR, however, needs a rough estimate of the concentration of
miRNAs of interest to carry out first , in order to make appropriate
dilutions; Non-template controls and a RT negative control must
be set up for each miRNA, when using a “primer pool method”
for retro-transcription; a chip-based dPCR method requires less
pipetting steps, which reduces potential PCR contamination, and
Quant StudioTM 3D chip has 20,000 fixed reaction wells, which
reduces variability of dPCR results (Figure 2 & 3 and Table 2 & 3).
To avoid bias and ensure that biomarker selection and outcome
assessment will not influence each other, a prospective specimen
collection retrospective blinded evaluation (PRoBE) design
randomized selection could be employed. An enrichment and
exfoliation strategy of colonocytes from stool for miRNA profiling
using Dynal superparamagnetic polystyrene beads coated with
a mouse IgG1 monoclonal antibody (Ab) Ber-Ep4, specific for an
epitope on the protein moiety of the glycopolypeptide membrane
antigen Ep-CAM, which is expressed on the surface of colonocytes
and colon carcinoma cells, can be used. Comparing the Agilent
electrophoretic (18S and 28S) patterns to those obtained from
total RNA extracted from stool, and differential lysis of colonocytes
by RT lysis buffer (Quagen), could be construed as a validation
that the electrophoretic pattern observed in stool (18S and 28S)
is truly due to the presence of human colonocytes, and not due to
stool contamination with Escherichia coli (16S and 23S). While
some exsosomal RNA can be released from purified colonocytes
into stool, correction for that effect can be made. Hence, for CRC
screening, miRNA markers are more comprehensive and preferable
to a DNA-, epigenetic-, mRNA- or a protein-based marker. An added
advantage of the use of the stable, nondegradable miRNAs by PCR
expression, or chip-based methods is being automatable, making
them more economical and acceptable by laboratory personnel
performing these assays.
To read more about this article....Open access Journal of Biostatistics & Biometric Applications
Please follow the URL to access more information about this article
https://irispublishers.com/abba/fulltext/absolute-dpcr-quantification-of-micrornas-by-absolute-dpcr-for-the-diagnostic-screening-of-colon-cancer.ID.000520.php