Authored by Wei-Liang Chen*,
Abstract
Hemimegalencephaly (HME) and focal cortical dysplasia type 2 (FCD2) are most common causes of childhood refractory epilepsy associated with severe developmental brain disorders, characterized by dyslamination, polymicrogyria, white matter heterotopia, immature neurons, calcifications, dysmorphic neurons (DN), and balloon cells (BC). Activating somatic and germline mutations of PI3K/AKT/mTOR signaling pathway have been identified in these phenotypes. Molecular diagnosis only can be reliably made via deep sequencing of affected brain tissue due to mosaicism. For those deemed not eligible for epilepsy surgery, establishing a molecular diagnosis can be challenging. Therefore, efforts have been taken to identify other biomarkers for these diagnoses. Phenotypes, including type of cortical malformation and other associated features, were found to highly correlate with the underlying genetic etiologies (genotype). Certain neurophysiologic features were also unique for these diagnoses. Furthermore, intracranial monitoring for epileptiform discharges could potentially impact the yield of molecular diagnostics. For example, level of mosaicism (variant allele frequency, VAF) was higher in region with more active interictal discharge seen in intraoperative electrocorticogram (ECoG). In this mini review, we will go over the current advance in molecular diagnostics for HME/FCD2, correlation between genotype and phenotype and future direction for expanding the role of neurophysiology in diagnosing these disorders.
Introduction
Hemimegalencephaly (HME) and focal cortical dysplasia type 2 (FCD2) are severe developmental brain disorders with shared neuropathologic and genetic features. Neuropathologic assessment of affected brain tissue shows common characteristic gross and microscopic changes, including large regions of dyslamination, polymicrogyria, white matter heterotopia, immature neurons, calcifications, dysmorphic neurons (DN), and balloon cells (BC). Germline (constitutional) and somatic (mosaic) mutations of critical genes in the phosphatidylinositol-3-kinase (PI3K)/AKT/mTOR signaling pathway – including PIK3CA, MTOR, AKT3, CCND2 and others – have been identified in those who with HME/FCD2. The presence of DNs and BCs is known to be highly correlated with epileptogenic foci. In fact, HME and FCD2 are the most common causes of childhood refractory epilepsy and often associated with poor neurodevelop mental outcomes [1,2]. Patients with HME are typically identified with onset of seizures in the first few months of life and age of seizure onset is usually before the age of one. Diagnosis is confirmed by MRI, wherein there are gross disruptions of at least three of the four cerebral lobes. Patients with FCD2 tend to have later onset seizure. Other neurological deficit and developmental issues are rare in FCD2. Seizure types associated with HME and FCD2 include infantile spasms, Ohtahara syndrome; myoclonic, tonic, and epilepsy partialis continua and other focal-onset seizures. Unfortunately, those seizure are typically resistant to pharmaceutical treatments. Cerebral hemispherectomy is the only current treatment for the intractable seizures associated with HME and more diffuse FCD2. However, more than 30% of children’s epilepsy cannot be controlled following surgical resection. Therefore, there is an urgent need of alternative treatments. As our knowledge of genomics becomes more advanced, pathway-specific drugs are becoming an appealing alternative [3]. Several targeted therapies including sirolimus (MTOR inhibitor), alpelisib (PI3K inhibitor), and miransertib (AKT inhibitor) are currently being examined for efficacy in vascular malformations and overgrowth in clinical trials [4] (clinicaltrials.gov: NCT03987152, NCT02638389, NCT04085653, NCT03094832) and the results will likely have clinical implication in HME/FCD managements. These drugs can impact therapeutics for those who do not respond to conventional treatments. However, in the majority of affected individuals, a molecular (genetic) diagnosis only can be made via deep sequencing of affected (or lesional) brain tissue due to genetic mosaicism. For those deemed not eligible for epilepsy surgery, establishing a molecular diagnosis can be challenging. Therefore, efforts have been taken to better correlate phenotype and genotype of this pathway.
Clinical phenotypes such as the degrees of malformation of HME and FCD2 are known to be related to causative genes and levels of mosaicism [3,5-7]. However, the seizure severity and the electroencephalogram (EEG) features (phenotype) to known genotypes is less clear. Our group recently works on systemically analyzing EEG data in patient with known HME/FCD2. This review paper aims to discuss our current understanding in molecular pathogenesis, and clinical and EEG phenotype/genotype correlation of this pathology.
Pathogenetic Mechanisms of HME and FCD
It will be helpful if we start with the definition of germline and somatic mutations. For the longest time, despite extensive efforts, the underlying genetic causes cannot be identified in patients with HME/FCD. Not until recently, we started to learn that somatic mutations found only in affected tissues (so called mosaicism) are difficult to be detected by sequencing the peripheral blood [8,9]. Sequencing peripheral blood, the traditional DNA source used in most clinical setting, is ideal for finding a variant exist in every single cell (so called germline “mutation” or pathogenic variant). However, for most of the somatic mutation that affects only parts of tissues (in this case, brain tissues), sequencing peripheral blood (leukocyte) barely has any yield. Thanks to the advance in molecular diagnostics for somatic mutations, causative genes of HME/FCD have been discovered recently. Using molecular inversion probe (mIP) capture technology targeting 10 genes involved in the PI3K/AKT/mTOR signaling pathway (PIK3CA, PIK3CB, PIK3CD, PIK3R1, PIK3R2, PIK3R3, AKT1, AKT2, AKT3 and PTEN), early study has shown that somatic mutations in affected brain tissues can be found in 12% of patient with HME [10]. For the first time, the key role that this pathway plays was unraveled. PI3K/AKT/mTOR signaling pathway is important in the integration of various environmental signals to regulate cell growth, proliferation, and metabolism [11]. Not surprisingly, hyperactivation of this pathway can be associated with cellular overgrowth seen in HME/FCD2. Later studies including more genes of PI3K/AKT/mTOR signaling pathway has identified more variants. In 41% of patient with FCD or HME, somatic mutations in AKT1, AKT3, MTOR, PIK3CA, TSC1 and TSC2 were found in resected brain tissue and germline pathogenic variant in DEPDC5, NPRL2, PIK3R2, TSC2 and MTOR in peripheral blood [8]. Furthermore, deeper sequencing (>2000 mean read depth) seems to have even higher yield to find causative mutations (in 59% of FCD cases and 87.5% of HME cases) [3].
Functionally, increased activation of this pathway was also evident. Phosphorylation of ribosomal protein S6 (pS6) is the marker of activation of PI3K/AKT/mTOR signaling pathway and was markedly increased in cells from the affected brain sections compared to non-HME cases [10]. Likewise, an increase in the level of phosphatidylinositol 3,4,5-triphosphate (PIP3), an indicator of PI3K activation, was shown in lymphoblastoid cells derived from individuals with megalencephaly compared with control cells [11]. Therefore, hyperactivation of PI3K/AKT/mTOR signaling pathway during neurodevelopment is believed to cause cellular hypertrophy observed in DNs and BCs. Indeed, more recent functional studies have suggested that mutations in genes of PI3K/AKT/mTOR signaling pathway alter differentiation, proliferation, and/or migration of neurons. For example, somatic mutations in AKT3 affecting neuronal progenitors limited in one cerebral hemisphere could lead to subsequent neuronal overgrowth and HME [12]. In addition, the somatic mutation in PI3K/AKT/mTOR signaling pathway is cell-type specific. Hyperactivation of this pathway restricted to the excitatory neuron lineage is sufficient to cause HME/FCD phenotypes, including cortical dyslamination, cytomegaly, DN and BC [8].
Although a substantial improvement in understanding the underlying molecular mechanisms of HME/FCD2 has been made, evidence still implies there is a more complex network. For example, even in tissues where no mutation detected, increased PI3K/ AKT/ mTOR pathway signaling and unique expression patterns remain evident, implicating other causative mechanisms/genes leading to a common pathophysiology. For instance, expressions of the T308 and S473 phosphorylated forms of AKT and in vitro AKT kinase activities were different between mutation-positive dysplasia cortex, mutation-negative dysplasia cortex, and non-dysplasia epilepsy cortex [10]. Making things even more complicated, a current study did not observe major differences among cases with distinct variants in MTOR or with AKT3, RHEB, or DEPDC5 variants. In fact, a less intense pS6 staining was consistently observed in more severely affected tissues from PIK- 3CA-HME subjects, reflecting possible complex feedback regulatory mechanisms occurring upon hyperactivation of this pathway [3] and a non-linear correlation between the biomarker and causative mutation. Nevertheless, as more and more patients have molecular diagnosis, our understanding about the clinical phenotype has progressed and correlation between genotype and phenotype has been unraveled.
Conclusion
Convalescent plasma is a promising safe and effective therapy for COVID-19. Without knowing antibody levels, particularly nAbs, data from the past clinical studies have prevented researchers from drawing definitive conclusions on the effectiveness of CPT. While further investigation on the timing of CP transfusion and CP units is important, stratification of CP recipients based on age, disease severity, comorbidities, or biomarkers will be critical for future clinical trials and successful application of CPT of COVID-19.
Correlation Between Genotype and Phenotype
Correlation between genotype and phenotype (geno-phenotype correlation) offer important information when it comes to molecular diagnostics. This is especially true for disorders caused by somatic mutations. Deeper sequencing has been proved to be necessary for detecting very low level of mosaicism from either blood sample [8] or affected tissues [3,13]. Therefore, in order to increase diagnostic yield, choosing limited number of genes with deeper coverage of sequencing is typically required. Better phenotyping always results in a stricter list of candidate genes and higher diagnostic yield.
The geno/phenotype correlation in HME/FCD2 has been recently explored by many groups. Microscopically, different genes can be associated with distinctive degrees of neuropathologic changes. For instances, somatic mutation of PIK3CA were associated with only mildly enlarged neuron, whereas mutations of AKT3 was associated with dramatically enlarged neuronal size [10]. Clinically, this correlation was also noticed. First of all, age of seizure onset seems to correlate well with the underlying genetic etiology. A current study found that a significant difference in the distribution of the age at seizure onset among groups of patients. Age of onset was earlier in patients with either PIK3CA (neonatal period), AKT3, or RHEB (before 6 months of age) variants, together with a poor cognitive outcome, compared to patients with somatic mutations in MTOR, TSC1/2, and SLC35A2 or germline variants in TSC2 or DEPDC5. This is also consistent with higher brain mosaicism rates (VAF from 7.5 to 34%) of variants found in PIK3CA, AKT3 and RHEB, reflecting an earlier timing of mutation occurrence during embryogenesis [3].
Furthermore, mutations of certain genes were only observed in given pathologies. For example, somatic loss of function mutations in SLC35A2 were only seen in FCD type I and mild cortical dysplasia, somatic MTOR mutation is mostly associated with FCD2 (19/20 patients), whereas PIK3CA is only seen in HME cases (4/4 patients) [3]. PIK3CA is also commonly associated with other more severe megalencephaly syndromes such as megalencepahly-capillary malformation syndrome (MCAP) but CNS-only phenotype like HME or dysplastic megalencephaly has been identified in the patient with hotspot somatic mutation as well [5]. It is likely determined by the distribution of the somatic mutation and the cell types involved. Nonetheless, non-CNS phenotypes sometimes can give a very informative clue about the underlying genotype.
Secondly, the types (somatic v.s. germline) of variants of each gene can influence the phenotype. For example, a previous study suggested that there are several distinct brain malformation syndromes caused by AKT3 mutations. The hotspot somatic AKT3 p.E17K mutation causes highly recognizable syndrome with megalencephaly, extensive focal cortical dysplasia (either HME or bilateral, multifocal FCD), and cutaneous vascular malformations. However, in patient with germline AKT3 pathogenic variants, three other distinctive phenotypes were seen i) megalencephaly-polymicrogyria with frequent asymmetry and occasionally patchy somatic findings; (ii) megalencephaly-polymicrogyria with periventricular nodular heterotopia; and (iii) megalencephaly with normal or minimal cortical dysplasia, and autistic features [6]. Similarly, somatic mutations in PIK3CA tend to cause more severe phenotype than germline pathogenic variants [9].
Lastly, when it comes to genotype, the somatic mutation is unique for its VAF (level of mosaicism). Not only mutation itself but also VAF of the mutation might plays important roles in determining phenotypes. They are most likely interactive as well. Early studies have suggested that higher mosaicism level more likely causes HME phenotype than lower mosaicism level, although there are exceptions and overlapping. For instances, one study showed that patients with HME has higher VAF (7.1-20.6%) than patient with FCD2 (2.3-10.6%) [8]. Conversely, some recent studies suggest a more important role of genes itself in determining phenotype. A recent study [3] with a larger cohort of FCD2 and HME patients showed that somatic MTOR mutation overwhelmingly cause FCD2 even with high VAF (as high as 18.6%). Amino acid position (codon) 2215 is the hotspot as p.S2215F and p.S2215Y are the most common somatic mutations, followed by p.A1459D. Similarly, another study showed that only one patient (1/20) who has p.A1459D mutation presented HME, rest of the MTOR mutations are associated with FCD2 phenotype, even in patients who has high VAF (15.7- 18.6% of p.L1460P and 6.5-8.4% of p.S2215F) [14]. These conflicting results might reflect one of the major obstacles for studying somatic mutation, the sampling bias. Sampling the most adequate and yielding location for genetic testing is often a “lucky dip”. Imagine that the neuropathologic features of the affect brain does not tightly correlate with the level of mosaicism. A researcher could have the anatomically most affected tissues for molecular diagnosis yet there is no guarantee these tissues are genetically most severe. Indeed, even in tissues with clear FCD2a pathology, mutation can still be missed [14].
Therefore, it boils down to another crucial question: could there be a marker other than neuropathology to guide which tissues/regions the molecular test should target? Some experts believe EEG might be the answer.
The Neurophysiologic (EEG) Findings of HME/FCD
Could the EEG help to distinguish genetically affected from unaffected tissues? Large-scale study is lacking. However, early intracranial data has revealed some convincing results. For example, a previous research [14] has found a higher pS6 level around the epileptogenic zone (identified by stereo EEG) in one patient who needed a repeat resection. This patient initially underwent temporal and occipital lobectomy with intraoperative electrocorticography performed at 7 months, followed by a staged additional craniotomy for left parietal resection with electrocorticography (ECoG) at 5 years of age, with resolution of epilepsy after the second procedure. Western blots from highly epileptogenic posterior temporal- occipital cortex resected during the first surgery had higher pS6 expression than parietal cortex resected during the second surgery. Intracranial EEG by depth electrodes and grids before the second surgery revealed seizure onset from the mesial parietal region, which exhibited elevated pS6 levels, but not from the lateral parietal region, which exhibited low pS6 expression. In another case in the same study, the VAF varied from 0 to 0.086 and demonstrated a striking gradient with higher VAF around the epileptogenic zone in the posterior temporal lobe and lower VAF at the periphery. Similar finding for the VAF gradient was also suggested by a more recent study utilizing targeted deep sequencing [3].
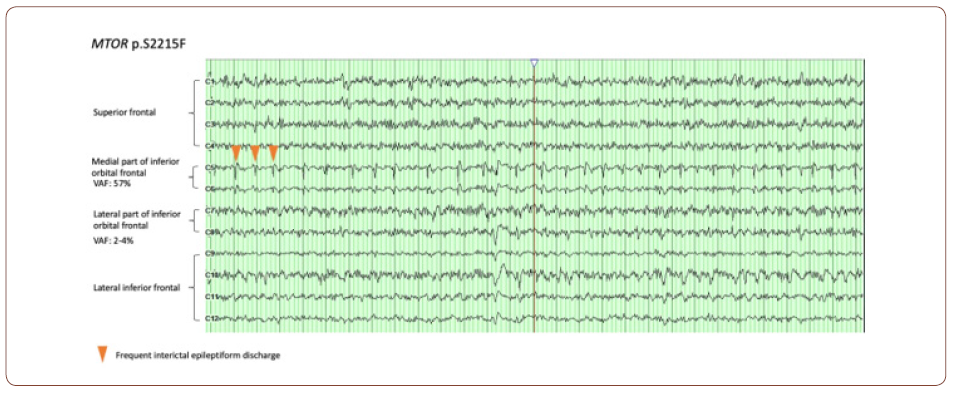
Therefore, the mutation itself seems to have a direct impact on EEG features. It has been noticed that some EEG features are enriched in certain genetic etiologies. For example, a higher occurrence of infantile spasms was observed in patients with SLC35A2 somatic variants (4/4 subjects) [3]. However, a more clinically relevant question is how strongly the spatial correlation between the presence of the mutation and epileptiform discharges. One way to answer this question is to put a dense array of electrodes over the abnormal cortices and systemically correlate the intracranial EEG data (e.g. epileptiform discharge) with VAF gradient among different parts of brain. This seemingly straightforward approach in fact could encounter many difficult practicability and ethical issues. First of all, putting a dense electrode grid on the surface of brain itself a major surgery. It is barely, if not never, indicated in such type of pathology given the EEG information unlikely change the management, especially for patients with HME or diffuse FCD requiring hemispherectomy. Secondly, even in patients with more focal pathology and EEG findings, resecting the unaffected brain tissues for genetic testing is ethically unjustifiable. Luckily for a relative larger lesion, it is still possible to compare the VAF gradient within lesion with intracranial EEG information. For example, electrocorticogram (ECoG) is a common procedure during surgery to assess the interictal discharges over the lesions. It is typically used to differentiate the electrically affected and unaffected cortices to help to determine resection margin. Therefore, ECoG can be an ideal tool for neurophysiologic pheno/genotype correlation. Indeed, we were able to correlate the VAF to the severity of interictal abnormality in some of our patients (see fig. for an example). An almost continuously run of epileptiform discharge (arrowhead) was seen in the region (medial part of inferior orbital frontal) where the VAF of MTOR mutation was highest (57%), whereas in the regions with less active discharge, the VAF is much lower (2-4% in lateral part of inferior orbital frontal).
Second important question is whether EEG features have any diagnostic and prognostic values. Again, large-scale systemic analysis of EEG in patients with HME/FCD is lacking. Nevertheless, one earlier study in 12 patients with HME showed three main types of EEG abnormalities in this cohort [15]:
1. Triphasic complexes: small negative wave followed by a large positive slow wave
2. Asymmetric burst suppression pattern: burst of alpha-like activity interrupted by hypoactive phases on affected side. Burst of large polymorphic spikes were seen on unaffected side
3. Alpha-like activity: asymmetric al and large-amplitude, sharp and non-reactive 7-12 Hz rhythm
In this cohort, all seizures were partial and 3 had infantile spasm. Two of them became seizure free after steroid treatment. Rest of them have seizure ranging from once a week to several times per day. Onset between 11 hours of age to 9 months of age. It seems that patient with alpha-like activity tend to have later seizure onset and longer survival, whereas in patients with triphasic complexes, onset is several days to weeks, poor developmental outcome and shorter life. One patient showed transition from triphasic complexes to alpha-like activity. This patient has better developmental outcome than patients presented with persistent triphasic complexes. Patients with burst-suppression pattern in this cohort responded to steroid well and had almost normal developmental outcome. However, none of those patients has genetic diagnosis. It is also unclear in this study whether the EEG findings were correlated with the anatomy and the severity of malformations.
A more recent study [16] included 9 patients with HME, 10 patients with FCD and 3 other types of malformations who underwent hemispherectomies partially unrevealed this question. Not surprisingly, among 9 patients with HME, about half of them (5/9) has focal EEG findings consistent with the underlying hemispheric abnormality. Seven patients with FCD also have focal spikes consistent with underlying focality. Unfortunately, in-detail description or EEG tracing are not available in this paper, which make systemic analysis and comparison become impossible. Nonetheless, these findings suggest that there is an association between the EEG and the underlying anatomical focality. Moreover, none of the patients in these studies done before the genomic era has molecular diagnoses to correlate with EEG findings. Therefore, further research is needed to elucidate the correlation between EEG features and molecular diagnoses. The answer will be important for those children who are not eligible for surgical resection yet need pathway-specific medicine for refractory epilepsy.
Conclusion
In conclusion, our understanding about molecular pathogenesis of HME/FCD2, the major causes for refractory epilepsy in pediatric population, has been improving substantially in the past few years. Correlation between the phenotype and genotype has been well described by many groups. It is still unclear if this correlation has prognostic value when it comes to the seizure outcome. A previous study showed no significant difference in surgery outcomes whether a mutation was detected or not [3]. This gap between genotype and clinical outcome reflects the fact that there is a more complex underlying network. Further study in neurophysiologic features of HME/FCD2 is needed for better understanding the underlying network.
To read more about this article...Open access Journal of Biology & Life Sciences
Please follow the URL to access more information about this article
To know more about our Journals...Iris Publishers





No comments:
Post a Comment